 |
 |
- Search
| Precis Future Med > Volume 2(1); 2018 > Article |
|
Abstract
Purpose
Addition of molecular markers to the American Joint Committee on Cancer (AJCC) staging system would allow further refinements in predicting recurrence and help individualize treatment decisions. We aimed to validate the Gastric Cancer Prognostic Score (GCPS) in an independent cohort using an easy and cost effective quantitative real-time polymerase chain reaction (qRT-PCR) assay.
Methods
We performed qRT-PCR using 48 samples from our previous study and expanded to 128 independent patients. The GCPS was recalculated using Cox regression estimates and the performance of cutoff values for GCPS was reassessed.
Results
The qRT-PCR results showed a similar pattern to nanostring data by scale function data comparison. Using a new cutoff value, GCPS stratified 95 stage IB–III patients who received adjuvant chemotherapy into 74 high-risk patients and 21 low-risk patients with significantly different recurrence-free survival (P< 0.0001). The survival difference remained significant (P= 0.028) in 27 patients who did not receive adjuvant chemotherapy. Among stage I and II patients who were treated with surgery only, one AJCC stage IIA patient was defined as low-risk and showed long-term survival. Nine of 12 high-risk patients showed recurrence less than 67 months after operation.
Although the incidence of gastric cancer (GC) has been declining for several decades, it is still the fifth most common cancer and the second most frequent (8.8%) cause of cancer death in the world [1]. The prognosis of GC varies considerably depending on tumor and patient characteristics [2]. Based on the American Joint Committee on Cancer (AJCC) Cancer Staging Manual, 7th Edition (2010), the AJCC tumor, node, metastasis (TNM) classification and staging system for gastric cancer was released. The outcomes of patients after curative surgery for advanced gastric cancer depend on pathologically diagnosed TNM stage, allowing physicians to develop a prognosis and design a treatment plan for individual patients. Unlike early gastric carcinomas without lymph node metastasis (AJCC stage IA) or inoperable stage IV disease, which have an excellent or dismal prognosis, respectively, in most cases, stage Ib to III cancers are heterogeneous; the 5-year survival rate is 57.4% in stage IB, 45.5% in IIA, 32.8% in IIB, 19.8% in IIIA, and 9.2% in IIIB [2,3]. In Korea, the 5-year relative survival rates and 95% confidence intervals (CIs) for localized, regional and distant metastatic gastric cancer are 92.4% (95% CI, 92.0 to 92.8), 55.7% (95% CI, 55.0 to 56.4), and 5.5% (95% CI, 5.0 to 6.0), respectively [4]. The addition of prognostic systems or molecular markers to the AJCC staging system would allow further refinements in predicting recurrence risk and might be helpful for individualized treatment decisions and postoperative counseling.
Recently, we reported a prognostic algorithm for gastric cancer—the Gastric Cancer Prognostic Score (GCPS)—comprised of eight genes (LAMP5, CDC25B, CDK1, CLIP4, LTB4R2, MATN3, NOX4, and TFDP1) that were discovered and validated in more than 700 stage II gastric cancer patients patients by gene expression profiling of archived paraffin-embedded tumor blocks [5]. In this study, we found that the GCPS identified GC patients at high-risk for recurrence regardless of adjuvant treatment, and that high-risk stage II GC patients showed similar disease-free survival to stage III patients regardless of specific Lauren type (diffuse or intestinal), warranting future validation with a fast, easy, and cost effective method using tissues from an outside hospital. We further validated the GCPS in a separate cohort using a quantitative real-time polymerase chain reaction (qRT-PCR) technique.
The independent cohort included 134 patients who received D2 gastrectomy for gastric cancer in Ajou University Hospital from January 1, 2005, to December 31, 2016, and included 93 males (69.4%). We were able to extract good quality RNA from archival tissue samples and obtain reliable qRT-PCR results in 128 of these gastric cancer patients.
For isolation of total RNA from formalin-fixed paraffin-embedded tissue, 10 (4-μm-thick) sections were cut from each tissue block. RNA was isolated using the RNeasy FFPE kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. These preparations were deparaffinized, followed by proteinase K treatment, on-column DNase digestion, and elution using Rnase-free water as described in the manufacturer’s protocol. Total RNA samples were stored at –80°C until use. RNA concentrations were measured using NanoDrop (Thermo Fisher Scientific, Wilmington, DE, USA).
Total RNA from each sample was reverse transcribed with the SuperScript VILO complementary DNA (cDNA) synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s recommendations. Target RNA (2 μg) was reverse transcribed using 5X VILO Reaction Mix and 10X SuperScript Enzyme Mix. cDNA synthesis was performed with the following thermal cycling parameters: 10 minutes at 25°C, 60 minutes at 42°C, and 5 minutes at 85°C (Applied Biosystems, Foster City, CA, USA). Finally, cDNA was diluted 1:5 prior to use in qRT-PCR .
Primers and TaqMan probes were designed using Primer Express (http://primer-express.software.informer.com/3.0/). The TaqMan probes were 5΄-labeled with the reporter fluorescent dye 6-carboxy-fluorescein (FAM), and 3΄ minor groove binder-nonfluorescent quencher (MGB-NFQ). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (VIC-MGB) was used as an endogenous control. All probes were purified by high-performance liquid chromatography.
PCR was performed on a 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) in 384-well plates using a final volume of 10 μL consisting of 5 μL 2X Taqman PreAmp Master Mix, 4 μL cDNA sample, and 1 μL primer/probe. PCR amplifications were performed in triplicate wells using the following conditions: 2 minutes at 50°C and 10 minutes at 94°C, followed by 40 two-temperature cycles of 95°C for 15 seconds and 60°C for 60 seconds. Differences in expression were determined by the relative quantification method, and the cycle threshold (Ct) values of the test genes were normalized to the Ct values of endogenous control GAPDH. The fold change was calculated using the equation 2-ΔCt.
In a previous study, we developed the GCPS using an n-Counter–based assay. For the GCPS prediction model, a gradient lasso algorithm was used. Using a cutoff of 0.2205 for GCPS in the n-Counter–based assay, gastric cancer patients were robustly divided into two groups: high-risk and low-risk. From the validation cohort of 216 patients treated with chemoradiotherapy in our previous study [5], we selected 48 samples for the qRT-PCR assay, 24 high-risk and 24 low-risk patients. The data were normalized using an internal reference gene, GAPDH. GCPS for each sample was calculated using Cox regression estimates that were defined in the previous study (Table 1). Kaplan-Meier survival curves were performed to assess performance of the cutoff value for GCPS.
The qRT-PCR results from 48 samples from previous study cohort [5] showed a similar pattern to our nanostring data by scale function data comparison (Fig. 1A). First, we chose 0.4424 as the cutoff point that divides the samples in half. Next, 50% of the samples were reclassified as high-risk and low-risk groups with the new cutoff point. Kaplan-Meier survival curves showed significantly different survival between low-risk and high-risk patients stratified by a new qRT-PCR results (P= 0.016) (Fig. 1B).
We obtained reliable qRT-PCR results in 128 cases, which were further analyzed with predefined algorithms. For 128 gastric cancer patients with AJCC stage IB–IV, the median age was 60 years (range, 27 to 80 years), and 13 cases (10.2%) were positive for Epstein-Barr virus. By Lauren classification, intestinal type was observed in 91 cases, diffuse in 28, and mixed in nine cases. Most of the patients (n= 101, 78.9%) received postoperative adjuvant chemotherapy. Recurrence occurred in 39 patients, and postoperative overall survival ranged from 1.9 to 71.6 months. AJCC stages included 16 patients with stage IB, 36 with stage IIA, 19 with stage IIB, 26 with stage IIIA, 24 with stage IIIB, and seven with stage IV disease. The clinicopathologic characteristics of 128 patients with or without recurrence are summarized in Table 2.
Using the methods described above, we defined 0.4424 as a new cutoff value from qRT-PCR results. As we observed a significant survival benefit of adjuvant chemotherapy (Fig. 2A), we divided the patient group according to the presence of adjuvant chemotherapy. Risk groups defined by GCPS using qRT-PCR stratified 95 stage IB–III patients who received adjuvant chemotherapy into 74 high-risk patients and 21 low-risk patients. The survival of these groups was statistically significantly different (P< 0.0001). In detail, out of 74 high-risk patients with stage IB–III, 52 died of disease, whereas only 15 of 21 low-risk patients in stage IB–III died of disease (Fig. 2B). Among stage IV patients, we identified four highrisk patients and two low-risk patients, but could not find any survival difference between them. The survival difference remained significant in 27 patients who did not receive adjuvant chemotherapy (P= 0.028) (Fig. 2C).
After subtraction of stage I and II patients without adjuvant chemotherapy, only one AJCC stage IIA patient was classified as low-risk, and this patient showed long-term recurrence-free survival after operation without chemotherapy. Among 12 (four IB, six IIA, two IIB in stage) patients defined as high-risk, nine (two IB, five IIA, two IIB in stage) died of disease within 67 months after operation (Fig. 2D).
To generate easy and cost effective molecular markers predicting recurrence in patients with gastric cancer, we validated and reproduced a previously described 8-gene based GCPS using a qRT-PCR assay with 48 tumor samples from the previous cohort and then further extended this to 128 gastric cancer tissues from an independent cohort at an outside hospital. After recalculating the qRT-PCR results, we developed a new cutoff value and validated the ability to predict recurrence in patients with AJCC stage IB to III gastric cancer.
Gastric cancer is the second most common cause of cancer-related death and is responsible for approximately one million deaths annually [6]. In Korea, gastric cancer is the most prevalent cancer in men and the second leading cause of cancer-related death [7]. The AJCC classification is based on depth of invasion, number of metastatic lymph nodes, and metastasis [8]. This system represents an excellent common language in the field of gastric cancer, but it does not include additional prognostic factors such as age, tumor size, or other tumor characteristics [9]. Given that some biological markers, such as oncogenes, tumor-suppressor genes, cell cycle regulators, and DNA repair genes, are related to tumorigenesis, growth, invasion, and metastasis, many investigators are searching for new predictive factors among these molecular markers [2]. Addition of such prognostic systems or molecular markers to the AJCC staging system would allow further refinements in predicting recurrence risk and can be helpful for individualized treatment decisions and postoperative counseling. In this aspect, our study is significant because we used the same gastric tumor samples with a different platform, reproduced similar results to the previous study, and then extended the study to a larger number of independent patients who underwent the same D2 gastrectomy at a different institute. Moreover, when we subtracted stage I and II patients without adjuvant chemotherapy, one patient classified as low-risk showed long-term recurrence-free survival after operation, whereas nine out of 12 patients defined as high-risk died of disease. This dramatic survival difference is similar to our previous results for gastric cancer patients treated with surgery only [5]. Our results imply that GCPS is a clinically applicable approach with low cost that can easily be performed in many institutes.
As recurrence is frequent among patients with gastric cancer after radical gastrectomy, it is important to stratify patients at a high-risk of recurrence. In our previous study [5], we clearly demonstrated that the presence of molecular heterogeneity in GC, which was associated with clinical outcomes but independent of clinicopathologic staging information. Our previous data indicated that stage IB/II patients had very poor prognosis when their tumors expressed high-risk gene signatures. Therefore, it might be necessary to prospectively design a trial to test whether chemoradiotherapy is required for stage IB/II patients with low-risk gene expression profiles. To minimize any potential bias from variations in clinical practice or surgery at a single center, we extended our patient cohort to 128 patients treated in an independent hospital during a similar period to our patient cohort. Moreover, to replace the original test that required special equipment and had high cost, we used almost the same algorithm with a different platform. Finally, we validated our GCPS results using the new platform.
In conclusion, we reproduced the 8-gene based GCPS with an easily applicable qRT-PCR assay and successfully predicted recurrence in patients with gastric cancer.
Fig. 1.
(A) The direct comparison of quantitative real-time polymerase chain reaction (qRT-PCR) and nanostring results showed a similar pattern by scale function data comparison. (B) Kaplan-Meier survival curves in 48 patients showed significantly different survival between low-risk and high-risk patients stratified by qRT-PCR results.
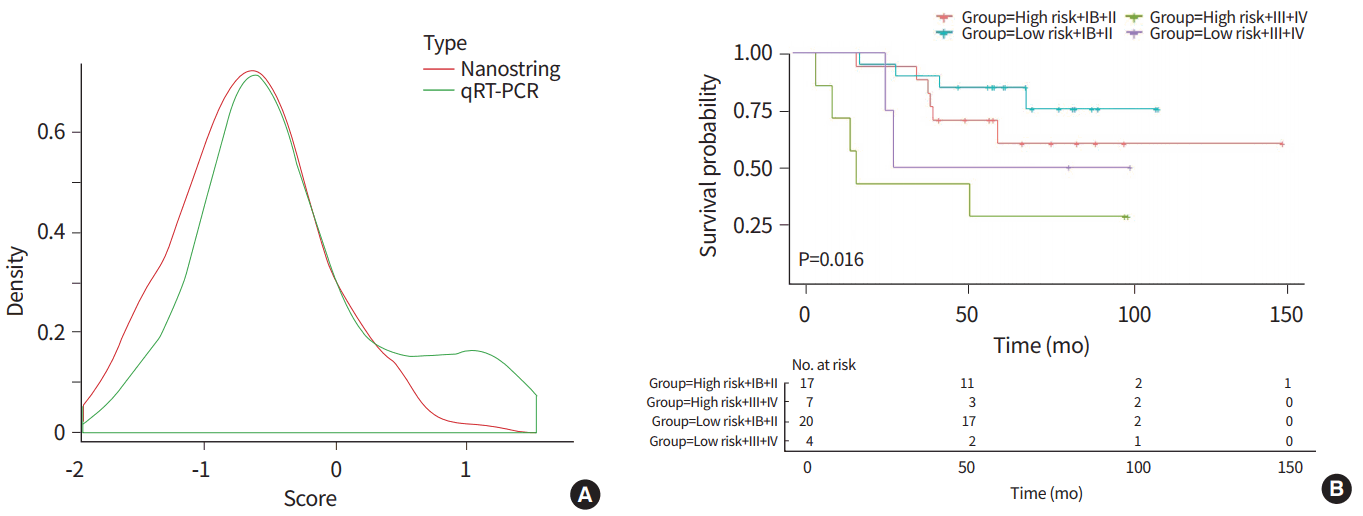
Fig. 2.
(A) In comparison of survival in patients with and without adjuvant chemotherapy, there was a significant survival benefit of adjuvant chemotherapy. (B) Stage IB–III patients who received adjuvant chemotherapy were stratified into 74 high-risk and 21 low-risk patients and the survival difference was statistically significantly different (P<0.0001). (C) Stage IB–III patients who did not receive adjuvant chemotherapy were stratified into 20 high-risk and 6 low-risk patients and the survival difference was statistically significantly different (P=0.028). (D) In stage I–II patients without adjuvant chemotherapy, one low-risk stage IIA patient showed long-term recurrence-free survival while nine of 12 high-risk patients were died of disease.
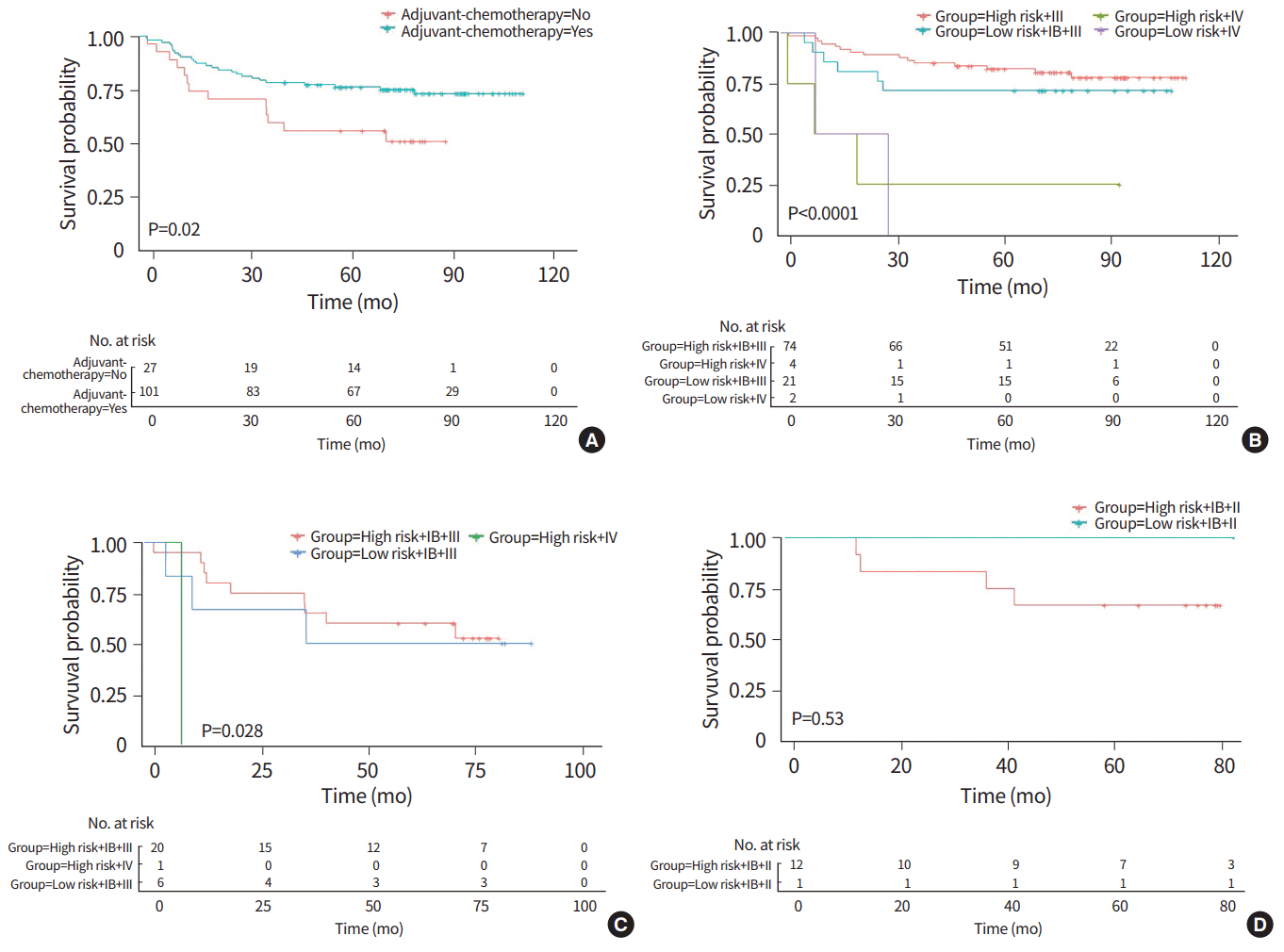
Table 1.
Cox regression estimate of genes used to calculate a new GCPS using a qRT-PCR assay
| Gene symbol | Cox regression estimate | |||
|---|---|---|---|---|
| LAMP5 | 0.0636 | |||
| CDC25B | -0.0175 | |||
| CDK1 | -0.1005 | |||
| CLIP4 | 0.4822 | |||
| LTB4R2 | -0.3950 | |||
| MATN3 | 0.2982 | |||
| NOX4 | 0.0288 | |||
| TFDP1 | -0.2886 |
Table 2.
Clinicopathologic characteristics of 128 patients with gastric cancer
REFERENCES
1. Muneoka Y, Akazawa K, Ishikawa T, Ichikawa H, Nashimoto A, Yabusaki H, et al. Nomogram for 5-year relapse-free survival of a patient with advanced gastric cancer after surgery. Int J Surg 2016;35:153–9.


2. Wu J, Liu X, Cai H, Wang Y. Prediction of tumor recurrence after curative resection in gastric carcinoma based on bcl-2 expression. World J Surg Oncol 2014;12:40.



3. Washington K. 7th Edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol 2010;17:3077–9.


4. Jung KW, Won YJ, Kong HJ, Oh CM, Shin A, Lee JS. Survival of Korean adult cancer patients by stage at diagnosis, 2006-2010: national cancer registry study. Cancer Res Treat 2013;45:162–71.



5. Lee J, Sohn I, Do IG, Kim KM, Park SH, Park JO, et al. Nanostring-based multigene assay to predict recurrence for gastric cancer patients after surgery. PLoS One 2014;9:e90133.



6. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006;24:2137–50.


8. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL. AJCC cancer staging manual. 7th ed. New York (NY): Springer; 2010.






