 |
 |
- Search
| Precis Future Med > Volume 5(3); 2021 > Article |
|
Abstract
Monogenic diabetes mellitus, which is diabetes caused by a defect in a single gene that is associated with β cell function or insulin action, accounts for 1% to 6% of all pediatric diabetes cases. Accurate diagnosis is important, as the effective treatment differs according to genetic etiology in some types of monogenic diabetes: high-dose sulfonylurea treatment in neonatal diabetes caused by activating mutations in KCNJ11 or ABCC8; low-dose sulfonylurea treatment in HNF1A/HNF4A-diabetes; and no treatment in GCK diabetes. Monogenic diabetes should be suspected by clinicians for certain combinations of clinical features and laboratory results, and approximately 80% of monogenic diabetes cases are misdiagnosed as type 1 diabetes or type 2 diabetes. Here, we outline the types of monogenic diabetes and the clinical implications of genetic diagnosis.
Diabetes mellitus (DM) is a heterogeneous metabolic disorder characterized by hyperglycemia resulting from inadequate insulin secretion, resistance to insulin action, or both [1]. The major clinical presentation of DM is elevated blood glucose concentration; however, insulin resistance and/or insulin deficiency also cause abnormal lipid and protein metabolism and mineral and electrolyte disturbance. These uncontrolled metabolic disturbances lead to various microand macrovascular complications that are associated with lower quality of life and increased risk of morbidity and mortality.
Most DM patients are classified as type 1 diabetes mellitus (T1DM) or type 2 diabetes mellitus (T2DM). T1DM is characterized by insulin deficiency caused by autoimmune β cell destruction with markers of autoimmune destruction at the time of diagnosis in approximately 90% of patients [2,3]. Autoimmune responses against β cells are triggered by environmental factors in genetically predisposed individuals. Many genetic loci, including human leukocyte antigen (HLA) complex as well as non-HLA genes including insulin (INS), protein tyrosine phosphatase non-receptor type 22 (PTPN22), cytotoxic T-lymphocyte associated protein 4 (CTLA4), are associated with T1DM with intermediate or low effect size [4]. Meanwhile, the key defects in T2DM are progressive dysfunction in β cell insulin secretion and insulin resistance [5,6]. T2DM is a multifactorial disease that is associated with environmental, epigenetic, and genetic risk factors. More than 40 genes have been identified to increase the risk of T2DM, although the effect sizes are small [5,7].
Approximately 1% to 6% of pediatric diabetes cases are caused by mutations in one of more than 40 genes that are associated with β cell function or insulin action and are termed monogenic diabetes [1,8]. The main categories of monogenic diabetes include neonatal diabetes mellitus (NDM), maturity-onset diabetes of the young (MODY), and syndromic diabetes. It is important that a precise etiologic classification is made in order to determine therapy, the prognosis of complications, and genetic diagnosis; however, misdiagnosis is frequent since the phenotypes are overlapped [8]. In this review, we introduce the types of monogenic diabetes, clinical implications of diagnosis, and strategies for approaching patients suspected of monogenic diabetes.
NDM is characterized by mild-to-severe hyperglycemia observed within 6 months of age that is associated with partial or complete insulin deficiency and can be classified as transient NDM (TNDM) or permanent NDM (PNDM) based on the clinical course [8]. The estimated incidence of NDM ranges from 1:90,000 to 1:260,000 among live births [9-11]. More than 30 genes have been identified in approximately 80% of NDM cases [12]. The common genetic causes and clinical features are summarized in Table 1. Clinical manifestations of NDM include hyperglycemia, glycosuria, polyuria, severe dehydration, and failure to thrive [13,14]. Also, infants with NDM are born small for gestational age or with intrauterine growth retardation, reflecting insulin deficiency in utero that underlines the importance of insulin in fetal growth.
PNDM that does not achieve significant treatment remission accounts for half of NDM cases. The majority of PNDM cases are caused by genes encoding KATP channel genes (potassium inwardly rectifying channel subfamily J member 11 [KCNJ11] and adenosine triphosphate [ATP] binding cassette subfamily C member 8 [ABCC8]) and genes encoding insulin itself (INS) [12]. The KATP channel consists of four subunits of the inward-rectifying potassium channel 6.2 (Kir6.2, encoded by KCNJ11) and four regulatory subunits of sulfonylurea receptor 1 (SUR1, encoded by ABCC8) and regulates insulin secretion in response to an increase in the ATP to adenosine diphosphate ratio, which makes the KATP channel close. The closure of the KATP channel leads to intracellular K+ accumulation and results in cell membrane depolarization, opening of voltage-gated Ca2+ channels, influx of Ca2+, and secretion of insulin [15]. Activating variants of KCNJ11 and ABCC8 keep the KATP channel in an open state regardless of ATP generation, leading to impaired insulin secretion [16]. Since KATP channels are expressed in the muscles and brain, approximately 20% of patients with KCNJ11 mutations are presented with muscular weakness, developmental delay, and early-onset epilepsy, in addition to NDM. Patients with KATP -NDM can be treated successfully with high-dose sulfonylureas, which are proven safe and effective for long-term glycemic control with only minimal hypoglycemia [17,18].
The second most common cause of PNDM is mutation in the preproinsulin gene (INS), which leads to misfolding proinsulin. The unfolded proinsulin results in loss of normal trafficking and accumulates in the endoplasmic reticulum (ER), leading to severe ER stress and apoptosis of β cells [19,20]. Two causative genes of MODY, GCK (encoding glucokinase) and Pancreatic And Duodenal Homeobox 1 (PDX1) (encoding the islet formation transcription factor) can be a rare cause of PNDM when in the homozygous or compound heterozygous state [21,22].
TNDM typically presents within a few weeks after birth and resolves spontaneously within 18 months of age, although it may relapse later in life [23]. Approximately 70% of TNDM cases are due to overexpression of the paternally imprinted genes at 6q24, including PLAG1 like zinc finger 1 (PLAGL1) and hydatidiform mole associated and imprinted (HYMAI), which results from paternally inherited duplication of 6q24, uniparental paternal disomy of the entire chromosome 6 or the segmental region that includes 6q24, or maternal hypomethylation of the differentially methylated region at 6q24 [14,24]. A transgenic mouse model of TNDM suggested that overexpression of PLAGL1/HYMAI, which has antiproliferative properties, leads to underdevelopment of the pancreas as well as impaired β cell function [25,26]. Compensatory increases in β cell number for reduced insulin synthesis and secretion restores normal fasting glucose, although hyperglycemia is sustained after the glucose challenge. Patients with 6q24-related TNDM are sensitive to insulin and achieve catch-up growth within a few weeks of treatment. Even after remission during infancy, it is important to be aware of the possibility of relapse around the time of adolescence [27].
The remaining causes of TNDM are mildly activating mutations in the KCNJ11 (Kir6.2) and ABCC8 (SUR1) genes, which encode KATP channels. In addition, it can, rarely, be caused by mutations in INS (encoding the preproinsulin), hepatocyte nuclear factor 1β (HNF1B; transcription factor), and solute carrier family 2 member 2 (SLC2A2; encoding the GLUT2 transporter) (encoding the GLUT2 transporter). These mutations are functionally less severe than those causing PNDM.
MODY is characterized by autosomal dominantly-inherited and non-autoimmune DM, which usually presents at a young age with some additional features. The estimated prevalence ranges from 1:10,000 to 1:23,000, varies by ethnic and racial group, between children and adults [28-30]. The majority of MODY cases are caused by three genes (GCK, hepatic nuclear factor 1 alpha [HNF1A], and HNF4A), although at least 14 genes have causative genes identified for MODY or a MODY-like phenotype (Table 2) [8].
Glucokinase, encoded by GCK, initiates glycolysis by facilitating the phosphorylation of glucose to glucose-6-phosphate and acts as a glucose sensor in the pancreatic β cell. The inactivation mutation of GCK increases the set point of glucosestimulated insulin secretion [31,32]. Most patients with GCK-MODY have mild fasting asymptomatic hyperglycemia within the prediabetic range and elevated hemoglobin A1c levels which usually do not exceed 7.5% [33,34]. GCK-MODY has a low risk of diabetes-related micro- and macrovascular complications, and treatment is not generally required in these patients except under pregnancy conditions [35,36]. An unaffected fetus born to a mother with GCK-MODY is associated with increased risk of the perinatal complications of gestational DM due to excessive insulin secretion in response to maternal hyperglycemia, while the affected fetus will have normal birth because an altered set point is shared. A fetus with paternally inherited GCK-MODY born to an unaffected mother is at risk for low birth weight [37].
The common symptomatic MODY that requires treatment is caused by mutations in HNF1A or HNF4A, which account for approximately 30% to 65% of all MODY cases [29,38,39]. These genes encode pancreatic transcription factors that are important for the transcription of genes related to β cell development and insulin secretion. The diagnosis of DM in patients with HNF1A/HNF4A-MODY is made in adolescence or young adulthood, related to the location of the mutations and exposure to maternal diabetes in utero [40-42]. Transient hyperinsulinemic hypoglycemia and large for gestational age can be observed in some patients with HNF1A/HNF4A-MODY [43,44]. The frequency of micro- and macrovascular complications is similar to that of patients with T1DM and T2DM [45]. Patients with HNF1A/HNF4A-MODY are sensitive to lowdose sulfonylureas and achieve better glycemic control than with insulin or metformin [46,47].
A syndromic DM should be considered in any child or adolescent with diabetes associated with multi-system extrapancreatic features [48]. The Online Mendelian Inheritance in Man website (www.ncbi.nlm.nih.gov/omim or www.omim.org) can help with clinical features to determine whether the gene for a particular syndrome has been defined and hence whether molecular genetic testing is available.
Syndromic NDM constitutes a relatively small fraction of NDM [12]. The spectrum of a genetic disorder can result in abnormalities in pancreas formation and function and may have accompanying severe congenital anomalies, such as intestinal and biliary atresia (regulatory factor X6 [RFX6]), congenital heart defects (GATA binding protein 6 [GATA6] or GATA binding protein 4 [GATA4]), brain malformations (pancreas associated transcription factor 1a [PTF1A], neuronal differentiation 1 [NEUROD1], immediate early response 3 interacting protein 1 [IER3IP1], motor neuron and pancreas homeobox 1 [MNX1], NK2 homeobox 2 [NKX2.2]), or skeletal dysplasia (eukaryotic translation initiation factor 2 alpha kinase 3 [EIF2AK3]), which are difficult to recognize during the neonatal period. In addition to mutations in forkhead box P3 (FOXP3), which is responsible for the IPEX syndrome (immune dysregulation, polyendocrinopathy, enteropathy, X-linked), several additional genes (signal transducer and activator of transcription 1 [STAT1], STAT3, LPS responsive beige-like anchor protein [LRBA], interleukin 2 receptor subunit alpha [IL2RA]) have now been described to have mutations causing infancy-onset syndromic diabetes, along with other autoimmune disorders that result from dysfunction of immune regulation [49-52].
Wolfram syndrome (WFS), also known as diabetes insipidus, DM, optic atrophy, and deafness syndrome, is diabetes associated with progressive optic atrophy below 16 years of age, although it may present anytime from early infancy [53]. Other typical clinical features including sensorineural deafness, central diabetes insipidus, urinary tract dysfunction, and neurological symptoms that develop later in life with variable order even within the same family [54-56]. At least 90% of patients have a recessive mutation in the WFS1 gene (Wolfram syndrome 1, WS1). Recently, a presentation similar to WS1 in many WFS1 mutation-negative patients was linked to a variant in CDGSH iron-sulfur domain 2 (CISD2) that is named Wolfram syndrome 2 (WS2). Clinical features of this variant do not include diabetes insipidus, instead, patients present with peptic ulcer bleeding and defective platelet aggregation [57].
Renal cysts and diabetes (RCAD) syndrome (HNF1B-MODY) is a rare subtype of familial diabetes, and heterozygous mutations in HNF1B rarely present with isolated diabetes [58]. Renal developmental disorders, mostly renal cysts and renal dysplasia, are present in almost all patients with HNF1B mutations or gene deletions even in the absence of diabetes [59,60]. Genital tract malformations, hyperuricemia, gout, and abnormal liver function test results can also occur [58]. Diabetes develops later, usually during adolescence or early adulthood, although TNDM has been reported in a few cases [61-64]. In addition to insulin deficiency related to pancreatic hypoplasia, patients also show some degree of hepatic insulin resistance and require early insulin therapy [65-67]. Also, lower exocrine pancreatic function with reduced fecal elastase presents in mutation carriers [68]. Therefore, the phenotype of RCAD patients is highly variable even within families sharing the same HNF1B mutation, and RCAD should be considered not only in the diabetes clinic but also in other clinics including nephrology, urology, and gynecology. In patients with renal cysts, imaging of the pancreas and fecal elastase should be considered. The absence of the pancreatic body and/or tail and abnormal fecal elastase indicates HNF1B-MODY [68]. One-third to two-thirds of cases showed de novo mutations and deletions of this gene [59,60].
Mitochondrial diabetes, also known as maternally inherited diabetes and deafness (MIDD), is caused by pathogenic variants in mitochondrial DNA, mostly tRNA variant m.3243A>G. Diabetes due to mitochondrial mutations and deletions is rarely seen in children and adolescents, and the majority of cases develop in young or middle-aged adults [69]. However, some cases have been reported in adolescents with a high degree of heteroplasmy [69-71]. Mitochondrial diabetes should be suspected in patients presenting with diabetes and sensorineural hearing loss inherited from the mother’s side or diabetes and progressive external ophthalmoplegia. Other clinical features such as macular pattern dystrophy, nephropathy, and neurological symptoms are more common in rarer forms of mitochondrial diabetes than classical forms [72]. Interestingly, the same m.3243A> G mutation also causes a much more severe clinical syndrome known as myopathy, encephalopathy, lactic acidosis, and stroke (MELAS) [73]. Patients with mitochondrial diabetes may respond initially to diet or oral hypoglycemic agents but often require insulin treatment within months or years. Metformin should be avoided as it interferes with mitochondrial function and may trigger episodes of lactic acidosis [74].
Genetic testing in patients suspected of monogenic diabetes has several clinical implications. Establishing a definite diagnosis is important for effective management based on the pathophysiology of diabetes. For example, avoiding insulin and sulfonylurea treatment can achieve successful glycemic control in the NDM caused by the activating mutations in KCNJ11 or ABCC8. Furthermore, genetic testing provides a better understanding of long-term prognosis and enables an avoidance of unnecessary treatment for diagnoses such a GCK-MODY, which has a low risk of diabetes-related complications and does not require treatment. Also, genetic diagnosis enables early evaluation and follow-up of extra-pancreatic manifestations of syndromic diabetes. For example, MODY caused by HNF1B mutations or 17q12 microdeletions is related to structural abnormalities in the kidney and urinary tract, hypomagnesemia, and abnormal liver function [68]. In the same way, DM caused by WFS1 has been associated with diabetes insipidus, optic atrophy, and deafness; CISD2 with optic atrophy, peptic ulcer bleeding, and defective platelet aggregation; and mitochondrial diabetes with deafness, nephropathy, and neurological symptoms [57,72,75].
There are limited practical guidelines for standardized diagnostic approaches and genetic testing for monogenic diabetes. Fig. 1 summarizes the proposed diagnostic approach for monogenic diabetes. The International Society for Pediatric and Adolescent Diabetes (ISPAD) has recommended genetic testing for patients diagnosed with DM before 6 months of age and patients diagnosed with DM between the age of 6 and 12 months without islet autoantibodies [8].
Approximately 80% of monogenic diabetes cases in pediatric and young adults are misdiagnosed as T1DM or T2DM; monogenic diabetes should be suspected by clinicians for patients with a certain combination of clinical and laboratory results [28,76]. Monogenic DM should be considered in T1DM patients who had negative autoantibodies at presentation, family history of diabetes in one parent, and preserved insulin secretion with detectable C-peptide at least 5 years after the presentation [8]. The features that should be suspected of monogenic diabetes in T2DM include lean body mass, lack of evidence of insulin resistance and/or other features of metabolic syndrome, family history of nonobese diabetes, and unusual fat distribution. Syndromic diabetes should be considered for DM cases with extra-pancreatic features such as deafness or neurological features.
As several causative genes have been identified in monogenic diabetes, the targeted gene panel, which consists of the genes considered to be associated with monogenic diabetes, is a rapid and cost-effective test. This targeted approach simplifies the interpretation of the results and minimizes the potential detection of secondary and incidental findings. Chromosomal microarray analysis also could be considered in patients with distinct features of a contiguous gene deletion such as 17q12 deletion syndrome. If the genetic cause is not detected with the targeted panel test, whole-exome sequencing (WES) and whole-genome sequencing (WGS) could be considered. WES and WGS are relatively unbiased genome-wide tests that cover protein-coding regions. WES and WGS allow for the discovery of new causative genes of monogenic diabetes. However, there is an issue of interpretation of results that include a large number of sequence variations and incidental findings. According to the recommendations in the American College of Medical Genetics and Genomics guidelines, the variants are assessed using various types of evidence and are classified into one of five categories: benign, likely benign, variant of uncertain significance, likely pathogenic, and pathogenic [77]. Incidental findings that are important to health and are medically actionable, such as those of cancer or cardiomyopathy, should be assessed and reported in patients undergoing WES or WGS [78].
Appropriate genetic counseling should be provided, and documentation of consent should be obtained when genetic testing is required [79]. This discussion contains the basic explanation of genetic disease and testing modalities; expected testing results; the advantages, disadvantages, risks, and limitations of testing; and testing cost. The possibility of incidental findings of medically actionable genes should be informed. Genetic counseling should be also provided after testing. The interpretation of test results, inheritance pattern, and risk of recurrence should be included. Also, information on available management, long-term prognosis, the need for further evaluation, and implications for the family should be provided.
Definite genetic diagnosis of monogenic diabetes is important for optimized treatment, notably in patients requiring no treatment (GCK-MODY), switching from insulin or metformin to sulfonylurea (HNF1A-MODY and HNF4A-MODY), and switching from insulin to high-dose sulfonylurea (KATP-NDM). In addition, a definite genetic diagnosis provides a better understanding of clinical course and familial recurrence risk. With recent advances in genetic testing technology, the capacity of genetic diagnosis has been increased with decreased cost. It is important for clinicians to know when and how to apply genetic testing in patients with DM. Therefore, careful, precise, and comprehensive assessment by clinicians is essential for suspicion of monogenic diabetes, encompassing patient and family history, physical examination, biochemical testing, and radiologic testing.
Notes
AUTHOR CONTRIBUTIONS
Conception or design: EK, YJR.
Acquisition, analysis, or interpretation of data: EK, LYC, YJK, KEO.
Drafting the work or revising: EK, LYC.
Final approval of the manuscript: EK, LYC, YJK, KEO, YJR.
Fig. 1.
Approach to genetic testing in patients with suspected monogenic diabetes. MODY, maturity-onset diabetes of the young; VUS, variants of uncertain significance; WES, whole exome sequencing.
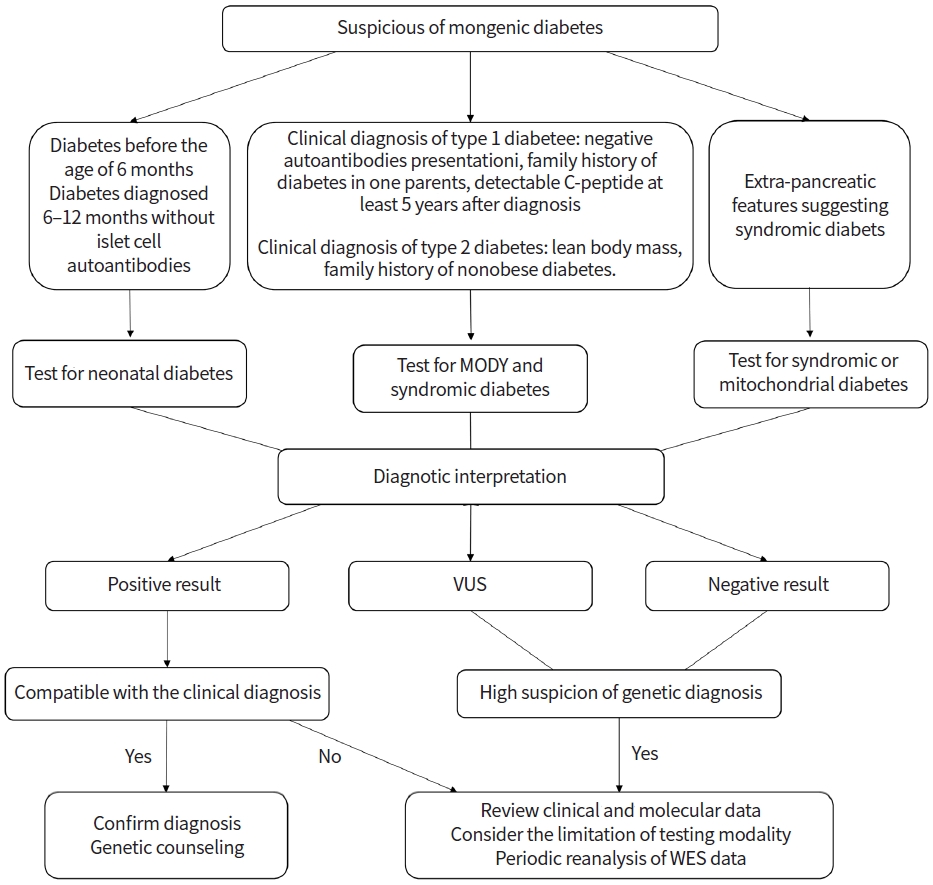
Table 1.
Genetic causes and clinical features of neonatal diabetes mellitus
Table 2.
Genetic causes and clinical implications of maturity-onset diabetes of the young
| Gene | Inheritance | Pathophysiology | Age of onset | Clinical implications |
|---|---|---|---|---|
| HNF4A | AD | Loss of function mutations of β cell transcription factor, disrupting β cell development and function | Preadolescence to young adulthood | Usually responds to low-dose sulfonylureas |
| GCK | AD or AR in NDM | Impaired β cell glucose sensing | Birth | Monitoring without treatment is possible |
| HNF1A | AD | Loss of function mutations of β cell transcription factor | Preadolescence to young adulthood | Usually responds to low-dose sulfonylureas |
| PDX1 | AR | Loss of function mutations of β cell transcription factor | Young adulthood | Treated with oral hypoglycemic agents or insulin, pancreatic agenesis in homozygotes/compound heterozygotes (rare) |
| HNF1B | AD | Loss of function mutations pancreatic/renal transcription factor | Preadolescence to young adulthood | Insulin treatment needed but genetic diagnosis allows monitoring of vital organs affected with gene defects such as renal structural abnormalities, genital tract malformations, pancreatic hypoplasia, hypomagnesemia, abnormal liver function, intellectual disabilities |
| NEUROD1 | AD | Loss of function mutations of β cell transcription factor | Young adulthood | Reduced penetrance for diabetes can be expected |
| KLF11a) | AD | Loss of function mutations of β cell transcription factor | Young adulthood | Treated with insulin |
| CEL | Deletion of variable number of tandem repeats | Pancreatic endocrine and exocrine dysfunction | Young adulthood | Insulin treatment needed but genetic diagnosis allows monitoring of pancreatic exocrine dysfunction in childhood and diabetes in later adulthood with multiple pancreatic cysts |
| PAX4a) | AD | Loss of function mutations of β cell transcription factor | Young adulthood | Treated with oral hypoglycemic agents or insulin |
| INS | AD or AR in NDM | Insulin gene mutation | Childhood to young adulthood | Early intensive insulin regimen is possible |
| BLKa) | AD | Insulin secretion defect | Young adulthood | Treated with oral hypoglycemic agents or insulin |
| ABCC8 | AD | Activating mutations of KATP channels | Preadolescence to young adulthood | Usually responds to high-dose sulfonylureas and early diagnosis allows monitoring of neurological abnormalities |
| KCNJ11 | AD | Activating mutations of KATP channels | Childhood | Usually responds to high-dose sulfonylureas and early diagnosis allows monitoring of neurological abnormalities |
| APPL1 | AD | Insulin secretion defect | Young adulthood | Treated with oral hypoglycemic agents or insulin |
| RFX6 | AD | Loss of function mutations of β cell transcription factor | Varies | Treated with diet, insulin, or oral hypoglycemic agents, reduced penetrance for diabetes can be expected |
REFERENCES
1. Addendum. 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2021. Diabetes Care 2021;44(Suppl 1):S15–33.


2. Eisenbarth GS. Type I diabetes mellitus: a chronic autoimmune disease. N Engl J Med 1986;314:1360–8.


3. Wang J, Miao D, Babu S, Yu J, Barker J, Klingensmith G, et al. Prevalence of autoantibody-negative diabetes is not rare at all ages and increases with older age and obesity. J Clin Endocrinol Metab 2007;92:88–92.


4. Lee HS, Hwang JS. Genetic aspects of type 1 diabetes. Ann Pediatr Endocrinol Metab 2019;24:143–8.



5. Castorani V, Polidori N, Giannini C, Blasetti A, Chiarelli F. Insulin resistance and type 2 diabetes in children. Ann Pediatr Endocrinol Metab 2020;25:217–26.



6. Todd JN, Srinivasan S, Pollin TI. Advances in the genetics of youth-onset type 2 diabetes. Curr Diab Rep 2018;18:57.



7. Ashcroft FM, Rorsman P. Diabetes mellitus and the β cell: the last ten years. Cell 2012;148:1160–71.



8. Hattersley AT, Greeley SA, Polak M, Rubio-Cabezas O, Njolstad PR, Mlynarski W, et al. ISPAD clinical practice consensus guidelines 2018: the diagnosis and management of monogenic diabetes in children and adolescents. Pediatr Diabetes 2018;19 Suppl 27:47–63.


9. Iafusco D, Massa O, Pasquino B, Colombo C, Iughetti L, Bizzarri C, et al. Minimal incidence of neonatal/infancy onset diabetes in Italy is 1:90,000 live births. Acta Diabetol 2012;49:405–8.


10. Wiedemann B, Schober E, Waldhoer T, Koehle J, Flanagan SE, Mackay DJ, et al. Incidence of neonatal diabetes in Austria-calculation based on the Austrian Diabetes Register. Pediatr Diabetes 2010;11:18–23.


11. Slingerland AS, Shields BM, Flanagan SE, Bruining GJ, Noordam K, Gach A, et al. Referral rates for diagnostic testing support an incidence of permanent neonatal diabetes in three European countries of at least 1 in 260,000 live births. Diabetologia 2009;52:1683–5.



12. De Franco E, Flanagan SE, Houghton JA, Lango Allen H, Mackay DJ, Temple IK, et al. The effect of early, comprehensive genomic testing on clinical care in neonatal diabetes: an international cohort study. Lancet 2015;386:957–63.



13. Sperling MA. Neonatal diabetes mellitus: from understudy to center stage. Curr Opin Pediatr 2005;17:512–8.


14. Polak M, Cave H. Neonatal diabetes mellitus: a disease linked to multiple mechanisms. Orphanet J Rare Dis 2007;2:12.



15. Ashcroft FM. ATP-sensitive potassium channelopathies: focus on insulin secretion. J Clin Invest 2005;115:2047–58.



16. Ashcroft FM, Puljung MC, Vedovato N. Neonatal diabetes and the KATP channel: from mutation to therapy. Trends Endocrinol Metab 2017;28:377–87.



17. Rafiq M, Flanagan SE, Patch AM, Shields BM, Ellard S, Hattersley AT, et al. Effective treatment with oral sulfonylureas in patients with diabetes due to sulfonylurea receptor 1 (SUR1) mutations. Diabetes Care 2008;31:204–9.


18. Bowman P, Sulen A, Barbetti F, Beltrand J, Svalastoga P, Codner E, et al. Effectiveness and safety of long-term treatment with sulfonylureas in patients with neonatal diabetes due to KCNJ11 mutations: an international cohort study. Lancet Diabetes Endocrinol 2018;6:637–46.


19. Stoy J, Edghill EL, Flanagan SE, Ye H, Paz VP, Pluzhnikov A, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci U S A 2007;104:15040–4.



20. Shaikh AA, Shirah B, Alzelaye S. A homozygous mutation in the insulin gene (INS) causing autosomal recessive neonatal diabetes in Saudi families. Ann Pediatr Endocrinol Metab 2020;25:42–5.



21. Njolstad PR, Sagen JV, Bjorkhaug L, Odili S, Shehadeh N, Bakry D, et al. Permanent neonatal diabetes caused by glucokinase deficiency: inborn error of the glucose-insulin signaling pathway. Diabetes 2003;52:2854–60.


22. De Franco E, Shaw-Smith C, Flanagan SE, Edghill EL, Wolf J, Otte V, et al. Biallelic PDX1 (insulin promoter factor 1) mutations causing neonatal diabetes without exocrine pancreatic insufficiency. Diabet Med 2013;30:e197–200.



23. Busiah K, Drunat S, Vaivre-Douret L, Bonnefond A, Simon A, Flechtner I, et al. Neuropsychological dysfunction and developmental defects associated with genetic changes in infants with neonatal diabetes mellitus: a prospective cohort study [corrected]. Lancet Diabetes Endocrinol 2013;1:199–207.


24. Temple IK, Shield JP. 6q24 Transient neonatal diabetes. Rev Endocr Metab Disord 2010;11:199–204.


25. Abdollahi A. LOT1 (ZAC1/PLAGL1) and its family members: mechanisms and functions. J Cell Physiol 2007;210:16–25.


26. Ma D, Shield JP, Dean W, Leclerc I, Knauf C, Burcelin R, et al. Impaired glucose homeostasis in transgenic mice expressing the human transient neonatal diabetes mellitus locus, TNDM. J Clin Invest 2004;114:339–48.



27. Docherty LE, Kabwama S, Lehmann A, Hawke E, Harrison L, Flanagan SE, et al. Clinical presentation of 6q24 transient neonatal diabetes mellitus (6q24 TNDM) and genotype-phenotype correlation in an international cohort of patients. Diabetologia 2013;56:758–62.


28. Shields BM, Hicks S, Shepherd MH, Colclough K, Hattersley AT, Ellard S. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia 2010;53:2504–8.


29. Pihoker C, Gilliam LK, Ellard S, Dabelea D, Davis C, Dolan LM, et al. Prevalence, characteristics and clinical diagnosis of maturity onset diabetes of the young due to mutations in HNF1A, HNF4A, and glucokinase: results from the SEARCH for Diabetes in Youth. J Clin Endocrinol Metab 2013;98:4055–62.



30. Shepherd M, Shields B, Hammersley S, Hudson M, McDonald TJ, Colclough K, et al. Systematic population screening, using biomarkers and genetic testing, identifies 2.5% of the U.K. pediatric diabetes population with monogenic diabetes. Diabetes Care 2016;39:1879–88.


31. Byrne MM, Sturis J, Clement K, Vionnet N, Pueyo ME, Stoffel M, et al. Insulin secretory abnormalities in subjects with hyperglycemia due to glucokinase mutations. J Clin Invest 1994;93:1120–30.



32. Osbak KK, Colclough K, Saint-Martin C, Beer NL, Bellanne-Chantelot C, Ellard S, et al. Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum Mutat 2009;30:1512–26.


33. Carmody D, Naylor RN, Bell CD, Berry S, Montgomery JT, Tadie EC, et al. GCK-MODY in the US National Monogenic Diabetes Registry: frequently misdiagnosed and unnecessarily treated. Acta Diabetol 2016;53:703–8.



34. Cascais M, Pereira E, Vieira A, Venancio M, Ramos L, Moleiro P. Hyperglycemia in pediatric age: could it be maturity onset diabetes of the young?: case reports and review of the literature. Ann Pediatr Endocrinol Metab 2019;24:262–6.



35. Steele AM, Shields BM, Wensley KJ, Colclough K, Ellard S, Hattersley AT. Prevalence of vascular complications among patients with glucokinase mutations and prolonged, mild hyperglycemia. JAMA 2014;311:279–86.


36. Stride A, Shields B, Gill-Carey O, Chakera AJ, Colclough K, Ellard S, et al. Cross-sectional and longitudinal studies suggest pharmacological treatment used in patients with glucokinase mutations does not alter glycaemia. Diabetologia 2014;57:54–6.


37. Hattersley AT, Beards F, Ballantyne E, Appleton M, Harvey R, Ellard S. Mutations in the glucokinase gene of the fetus result in reduced birth weight. Nat Genet 1998;19:268–70.


38. Yamagata K, Furuta H, Oda N, Kaisaki PJ, Menzel S, Cox NJ, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature 1996;384:458–60.


39. Yamagata K, Oda N, Kaisaki PJ, Menzel S, Furuta H, Vaxillaire M, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3). Nature 1996;384:455–8.


40. Stride A, Vaxillaire M, Tuomi T, Barbetti F, Njolstad PR, Hansen T, et al. The genetic abnormality in the beta cell determines the response to an oral glucose load. Diabetologia 2002;45:427–35.


41. Harries LW, Ellard S, Stride A, Morgan NG, Hattersley AT. Isomers of the TCF1 gene encoding hepatocyte nuclear factor-1 alpha show differential expression in the pancreas and define the relationship between mutation position and clinical phenotype in monogenic diabetes. Hum Mol Genet 2006;15:2216–24.


42. Bellanne-Chantelot C, Carette C, Riveline JP, Valero R, Gautier JF, Larger E, et al. The type and the position of HNF1A mutation modulate age at diagnosis of diabetes in patients with maturity-onset diabetes of the young (MODY)-3. Diabetes 2008;57:503–8.


43. Pearson ER, Boj SF, Steele AM, Barrett T, Stals K, Shield JP, et al. Macrosomia and hyperinsulinaemic hypoglycaemia in patients with heterozygous mutations in the HNF4A gene. PLoS Med 2007;4:e118.



44. Yau D, Colclough K, Natarajan A, Parikh R, Canham N, Didi M, et al. Congenital hyperinsulinism due to mutations in HNF1A. Eur J Med Genet 2020;63:103928.


45. Steele AM, Shields BM, Shepherd M, Ellard S, Hattersley AT, Pearson ER. Increased all-cause and cardiovascular mortality in monogenic diabetes as a result of mutations in the HNF1A gene. Diabet Med 2010;27:157–61.


46. Shepherd M, Shields B, Ellard S, Rubio-Cabezas O, Hattersley AT. A genetic diagnosis of HNF1A diabetes alters treatment and improves glycaemic control in the majority of insulin-treated patients. Diabet Med 2009;26:437–41.


47. Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 2003;362:1275–81.


48. Schmidt F, Kapellen TM, Wiegand S, Herbst A, Wolf J, Frohlich-Reiterer EE, et al. Diabetes mellitus in children and adolescents with genetic syndromes. Exp Clin Endocrinol Diabetes 2012;120:579–85.


49. Uzel G, Sampaio EP, Lawrence MG, Hsu AP, Hackett M, Dorsey MJ, et al. Dominant gain-of-function STAT1 mutations in FOXP3 wild-type immune dysregulation-polyendocrinopathy-enteropathy-X-linked-like syndrome. J Allergy Clin Immunol 2013;131:1611–23.



50. Flanagan SE, Haapaniemi E, Russell MA, Caswell R, Allen HL, De Franco E, et al. Activating germline mutations in STAT3 cause early-onset multi-organ autoimmune disease. Nat Genet 2014;46:812–4.



51. Johnson MB, De Franco E, Lango Allen H, Al Senani A, Elbarbary N, Siklar Z, et al. Recessively inherited LRBA mutations cause autoimmunity presenting as neonatal diabetes. Diabetes 2017;66:2316–22.


52. Roth TL, Puig-Saus C, Yu R, Shifrut E, Carnevale J, Li PJ, et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 2018;559:405–9.



53. Inoue H, Tanizawa Y, Wasson J, Behn P, Kalidas K, Bernal-Mizrachi E, et al. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat Genet 1998;20:143–8.


54. Marshall BA, Permutt MA, Paciorkowski AR, Hoekel J, Karzon R, Wasson J, et al. Phenotypic characteristics of early Wolfram syndrome. Orphanet J Rare Dis 2013;8:64.



55. Karzon R, Narayanan A, Chen L, Lieu JE, Hershey T. Longitudinal hearing loss in Wolfram syndrome. Orphanet J Rare Dis 2018;13:102.



56. Bueno GE, Ruiz-Castaneda D, Martinez JR, Munoz MR, Alascio PC. Natural history and clinical characteristics of 50 patients with Wolfram syndrome. Endocrine 2018;61:440–6.


57. Rigoli L, Di Bella C. Wolfram syndrome 1 and Wolfram syndrome 2. Curr Opin Pediatr 2012;24:512–7.


58. Bingham C, Hattersley AT. Renal cysts and diabetes syndrome resulting from mutations in hepatocyte nuclear factor-1beta. Nephrol Dial Transplant 2004;19:2703–8.

59. Bellanne-Chantelot C, Clauin S, Chauveau D, Collin P, Daumont M, Douillard C, et al. Large genomic rearrangements in the hepatocyte nuclear factor-1beta (TCF2) gene are the most frequent cause of maturity-onset diabetes of the young type 5. Diabetes 2005;54:3126–32.


60. Ulinski T, Lescure S, Beaufils S, Guigonis V, Decramer S, Morin D, et al. Renal phenotypes related to hepatocyte nuclear factor-1beta (TCF2) mutations in a pediatric cohort. J Am Soc Nephrol 2006;17:497–503.


61. Edghill EL, Bingham C, Ellard S, Hattersley AT. Mutations in hepatocyte nuclear factor-1beta and their related phenotypes. J Med Genet 2006;43:84–90.

62. Raile K, Klopocki E, Holder M, Wessel T, Galler A, Deiss D, et al. Expanded clinical spectrum in hepatocyte nuclear factor 1b-maturity-onset diabetes of the young. J Clin Endocrinol Metab 2009;94:2658–64.


63. Yorifuji T, Kurokawa K, Mamada M, Imai T, Kawai M, Nishi Y, et al. Neonatal diabetes mellitus and neonatal polycystic, dysplastic kidneys: Phenotypically discordant recurrence of a mutation in the hepatocyte nuclear factor-1beta gene due to germline mosaicism. J Clin Endocrinol Metab 2004;89:2905–8.

64. Edghill EL, Bingham C, Slingerland AS, Minton JA, Noordam C, Ellard S, et al. Hepatocyte nuclear factor-1 beta mutations cause neonatal diabetes and intrauterine growth retardation: support for a critical role of HNF-1beta in human pancreatic development. Diabet Med 2006;23:1301–6.

65. Bellanne-Chantelot C, Chauveau D, Gautier JF, Dubois-Laforgue D, Clauin S, Beaufils S, et al. Clinical spectrum associated with hepatocyte nuclear factor-1beta mutations. Ann Intern Med 2004;140:510–7.


66. Pearson ER, Badman MK, Lockwood CR, Clark PM, Ellard S, Bingham C, et al. Contrasting diabetes phenotypes associated with hepatocyte nuclear factor-1alpha and -1beta mutations. Diabetes Care 2004;27:1102–7.


67. Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab 2008;4:200–13.

68. Tjora E, Wathle G, Erchinger F, Engjom T, Molven A, Aksnes L, et al. Exocrine pancreatic function in hepatocyte nuclear factor 1β-maturity-onset diabetes of the young (HNF1B-MODY) is only moderately reduced: compensatory hypersecretion from a hypoplastic pancreas. Diabet Med 2013;30:946–55.


69. Reinauer C, Meissner T, Roden M, Thon A, Holterhus PM, Haberland H, et al. Low prevalence of patients with mitochondrial disease in the German/Austrian DPV diabetes registry. Eur J Pediatr 2016;175:613–22.


70. Guillausseau PJ, Dubois-Laforgue D, Massin P, Laloi-Michelin M, Bellanne-Chantelot C, Gin H, et al. Heterogeneity of diabetes phenotype in patients with 3243 bp mutation of mitochondrial DNA (maternally inherited diabetes and deafness or MIDD). Diabetes Metab 2004;30:181–6.


71. Laloi-Michelin M, Meas T, Ambonville C, Bellanne-Chantelot C, Beaufils S, Massin P, et al. The clinical variability of maternally inherited diabetes and deafness is associated with the degree of heteroplasmy in blood leukocytes. J Clin Endocrinol Metab 2009;94:3025–30.


72. Decoux-Poullot AG, Bannwarth S, Procaccio V, Lebre AS, Jardel C, Vialettes B, et al. Clinical phenotype of mitochondrial diabetes due to rare mitochondrial DNA mutations. Ann Endocrinol (Paris) 2020;81:68–77.


73. Goto Y, Nonaka I, Horai S. A mutation in the tRNA(Leu) (UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 1990;348:651–3.


74. Lalau JD. Lactic acidosis induced by metformin: incidence, management and prevention. Drug Saf 2010;33:727–40.

75. De Franco E, Flanagan SE, Yagi T, Abreu D, Mahadevan J, Johnson MB, et al. Dominant ER stress-inducing WFS1 mutations underlie a genetic syndrome of neonatal/infancy-onset diabetes, congenital sensorineural deafness, and congenital cataracts. Diabetes 2017;66:2044–53.



76. Donath X, Saint-Martin C, Dubois-Laforgue D, Rajasingham R, Mifsud F, Ciangura C, et al. Next-generation sequencing identifies monogenic diabetes in 16% of patients with late adolescence/adult-onset diabetes selected on a clinical basis: a cross-sectional analysis. BMC Med 2019;17:132.



77. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405–24.



-
METRICS

-
- 1 Crossref
- 3,515 View
- 127 Download






