Functional roles of heterogeneous nuclear ribonucleoprotein K in post-transcriptional gene regulation
Article information
Abstract
Since it is widely accepted that the accumulation of genetic alterations is the main cause of cancer, understanding how cancer-associated genes are regulated is crucial to the development of cancertherapies. As one of important RNA-binding proteins (RBPs), heterogeneous nuclearribonucleoprotein K (hnRNPK), is known to regulate the expression of target genes involved in various pathways, such as transcription, splicing, and translation. HnRNPK is also closely associated with cancer progression, including the acquisition of metastatic potential. At the post-transcriptional level, gene expression is determined by competitive or cooperative interactions betweentrans-acting factors including RBPs and non-coding RNAs (ncRNAs) which are capable of binding to cis-elements in target genes. In this review, we discuss the roles of hnRNPK in post-transcriptional gene regulation. The regulation of cancer-associated genes (oncogenes and tumor suppressors) via crosstalk between hnRNPK and ncRNAs such as microRNAs and long ncRNAs is described in detail. This review highlights how hnRNPK may be a promising targetforthe development of cancertherapeutics.
INTRODUCTION
At the RNA level, gene expression is controlled through post-transcriptional gene regulation (PTGR). PTGR is determined by the combination of cis-acting element(s) presented in target messenger RNA (mRNA) and trans-acting factor(s) that recognizes and binds to specific cis-acting sequences. As one group of the trans-acting factors, RNA-binding proteins (RBPs) have been accepted as key players in post-transcriptional events. Through sequence-specific interaction between RNA-binding domain and target mRNA, RBPs are closely associated with mRNA splicing, mRNA stability, or translation, thus making a huge impact on target gene expression. RBPs are known to have one or more RNA binding domains such as RNA recognition motif, K homology motif (KH), and arginine-glycine-glycine box. Emerging evidences indicate that RBP plays critical roles in cancer development via acceleration of cancer progression and promotion of cancer aggressiveness. Therefore, understanding the action mechanism of RBP has been expected to contribute to develop prognostic biomarkers and provide a new paradigm for cancer treatment. This review will focus on the oncogenic function of heterogeneous nuclear ribonucleoprotein K (hnRNPK) in terms of PTGR.
STRUCTURE AND LOCALIZATION OF hn-RNPK
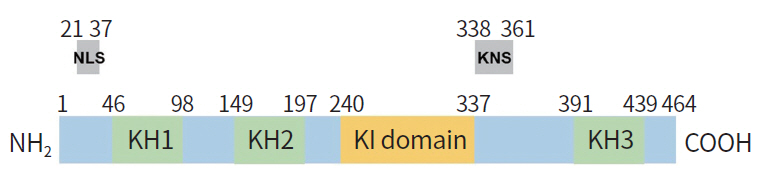
HnRNPK is an approximately 65 kDa protein mapped to chromosome 9 in humans [1]. It is a highly conserved RBP and is involved in multiple gene regulation processes. As a heterogeneous nuclear RNA binding protein (hnRNP), it is abundantly expressed in various human cells and localizes in both the nucleus and cytoplasm [2]. Each member of the hnRNP family exhibits different RNA binding motifs and specificities. Compared to other related proteins, hnRNPK preferentially recognizes poly-C sequences in the 3′ untranslated region (UTR) of target mRNA; it is able to interact with RNA or single-strand DNA through its three repeats of KH domains which consist of about 65 to 70 highly conserved amino acids (Fig. 1) [3,4]. Through yeast three-hybrid screening and computational analysis, KH domains were shown to be responsible for the interaction between hnRNPK and its target mRNAs [5]. Although three KH domains are reported to cooperatively function in hnRNPK-elicited gene regulation, no detailed mechanism of how KH domains bind to the target mRNA has been reported [6]. HnRNPK also contains a nuclear localization signal (NLS) and a nuclear shuttling domain (K nuclear shuttling [KNS]) which enables its translocation between the cytoplasm and the nucleus [7,8]. In addition, the K-protein–interactive (KI) region positioned between the KH2 and KH3 domains plays important roles in interacting with other proteins, including various tyrosine kinases, which suggests that hnRNPK is able to cross-talk with multiple signaling molecules [9].
FUNCTIONAL ROLES OF hnRNPK IN POST-TRANSCRIPTIONAL GENE REGULATION
Gene regulation is largely divided into two mechanisms: transcriptional and PTGR. HnRNPK has been reported to regulate gene expression by both mechanisms. Several target genes have been reported to be regulated at the transcriptional level, such as mu (μ) opioid receptor [10] and eukaryotic initiation factor 4E [11]. In this review, we focused on the functional roles of hnRNPK in PTGR. PTGR regulates gene expression at the RNA level by affecting RNA stability, splicing, transport, and translation (Fig. 2).
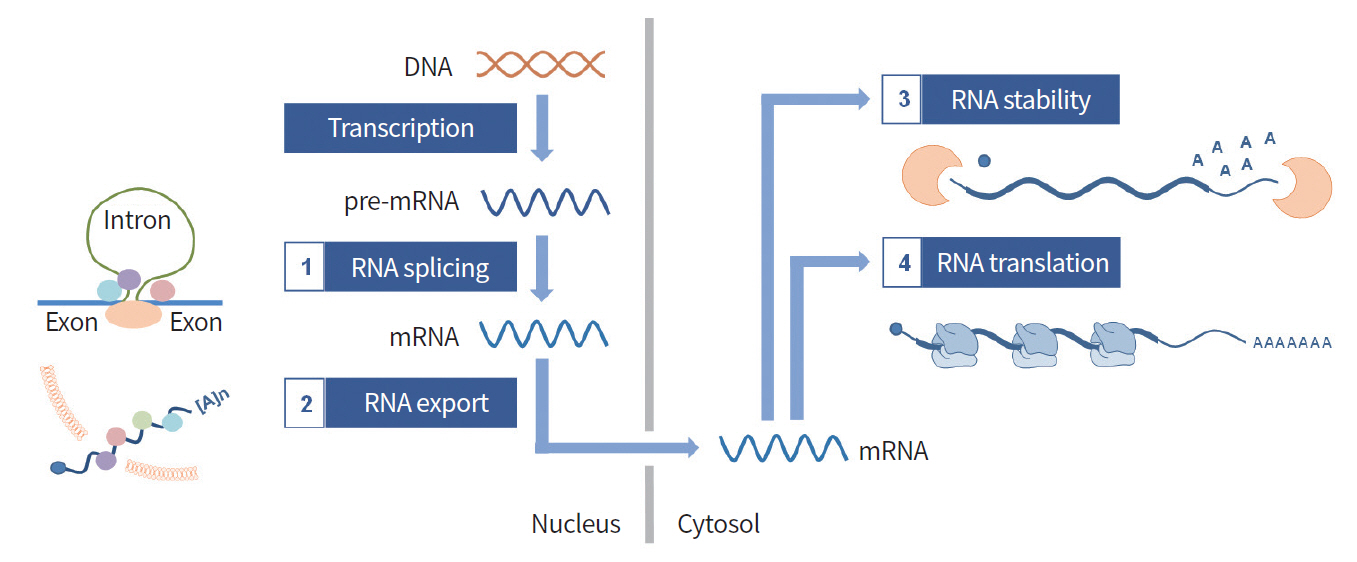
Four main steps of post-transcriptional gene regulation (PTGR). PTGR is the gene regulation at RNA level. After RNA is transcribed by RNA polymerase II, pre-mRNA undergoes splicing to be maturated. The export of mRNA is determined by many RNA-binding proteins. In the cytoplasm, gene expression is governed via mRNA stability and translation.
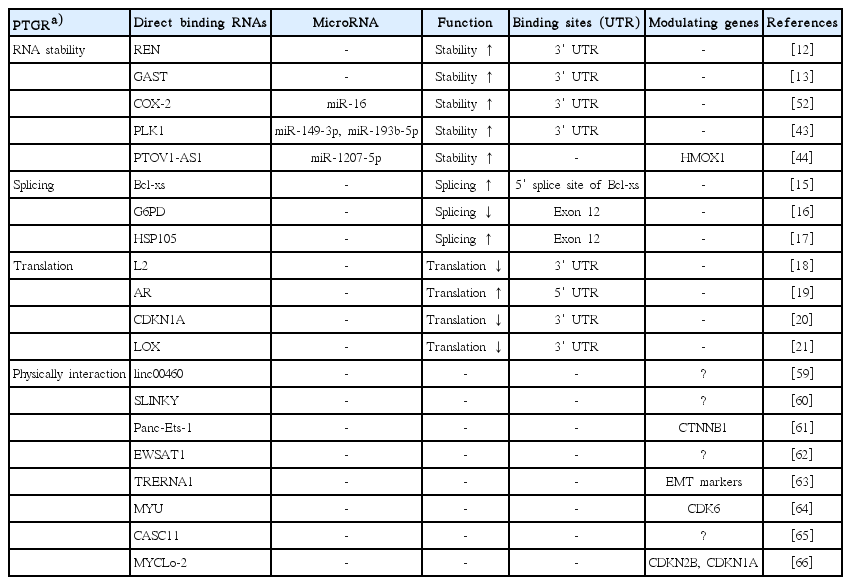
Several studies have reported that hnRNPK controls the expression of target genes by regulating the stability of their mRNA. Skalweit et al. [12] reported that renin (REN) mRNA is stabilized by hnRNPK and other four RNA binding proteins such as hnRNPE1, dynamin, Y-box binding protein 1 (YB1), and MINT-homologous protein. Along with poly (C) binding protein (PCBP1) and nucleolin, hnRNPK increases epidermal growth factor-mediated expression of the gastrointestinal hormone gastrin by stabilizing its mRNA [13]. By functioning as a splicing regulator, hnRNPK is also able to regulate the expression of its target genes. It has been shown that hnRNPK is closely associated with 50% of the alternative splicing processes that occur in apoptotic genes [14]. For example, hnRNPK can bind to the 5′ splice site of pro-apoptotic Bcl-2-like 1 (BCL2L1) small (BCLXS) mRNA and suppress its expression by interfering with splicing events [15]. Another evidence of hn-RNPK as a splicing regulator is the starvation-elicited splicing of glucose-6-phosphate dehydrogenase (G6PD) pre-mRNA. Ribonucleoprotein immunoprecipitation (RNP-IP) experiments showed that hnRNPK interacts with G6PD pre-mRNA, blocks its splicing, and decreases its expression [16]. In addition, hnRNPK is involved in heat stress-induced alternative splicing, such as exon 12 exclusion of heat shock protein family H member 1 (HSP105) pre-mRNA [17]. Translational gene regulation is also responsible for the function of hnRNPK. RNA gel shift assays showed that the poly-r (C) binding protein hnRNPK and PCBP interact with human papillomavirus type 16 L2 mRNA and inhibit its translation [18]. Depending on the location of UTRs containing the consensus binding sequences of hnRNPK, the translational efficiency of target mRNAs is differentially affected. For example, hnRNPK enhances the expression of androgen receptor by directly binding to the 5′UTR of its mRNA [19]. On the other hand, Yano et al. [20] demonstrated that translation of p21 mRNA is competitively regulated by HuB and hnRNPK. During late-stage erythropoiesis, hnRNPK is also able to regulate the expression of lysyl oxidase (LOX) mRNA by binding to the 3′UTR of its mRNA [21].
THE KEY ROLE OF hnRNPK IN CANCER
Several studies have demonstrated that hnRNPK is highly expressed in diverse types of cancer tissues as compared to its expression in corresponding normal tissues. Moreover, the expression level of hnRNPK is gradually increased with increasing tumor stage, suggesting that hnRNPK is closely linked to cancer progression including the acquisition of metastatic potential [22-34]. In addition, hnRNPK is dramatically abundant in the cytoplasm of neoplastic tissues as compared to that of non-neoplastic tissues. Increased levels of cytoplasmic hnRNPK are closely related with the prognosis of cancer patients [35,36]. Otoshi et al. [37] recently reported that hnRNPK is strongly expressed in renal cell carcinoma (RCC) and the cytoplasmic localization of hnRNPK is more increased in metastatic RCC than in non-metastatic RCC specimens. Cytoplasmic localization of hnRNPK is known to be associated with its phosphorylation status. Basically, phosphorylated hnRNPK at serine 284 and 353 by mitogen-activated protein kinase/extracellular-signal-regulated kinase (MAPK/ERK) leads its cytoplasmic accumulation, where it regulates target mRNA translation [38]. For these reasons, cytoplasmic accumulation of hnRNPK is known as an effective biomarker for cancer.
HnRNPK is also closely associated with apoptosis and cancer metastasis. Inoue et al. [39] reported that aberrant localization of hnRNPK in the cytoplasm is responsible for the acquisition of metastatic potential, including migratory and invasive abilities. Furthermore, they demonstrated that hnRNPK-overexpressed cells show characteristics of malignant cancer such as high invasiveness. By analyzing microarray data, they also identified that a group of genes that are involved in cell motility and angiogenesis are governed by hn-RNPK [40]. Cancer cells showing high hnRNPK expression are resistant to various stresses. Similarly, hnRNPK knockdown induces apoptotic cell death by activating caspases [41]. It has been also reported that many anti-apoptotic genes have been identified as hnRNPK targets including cyclin D1, G0/G1 switch2, X-linked inhibitor of apoptosis (XIAP)-associated factor 1, and ERCC excision repair 4, endonuclease catalytic subunit (ERCC4) [41,42]. Our recent studies also demonstrated that hnRNPK knockdown not only reduces cell viability and colony-forming ability but also increases poly (ADP-ribose) polymerase (PARP) cleavage [43,44]. In summary, hnRNPK has a critical role for cancer cell progression and metastasis.
INTERPLAY BETWEEN hnRNPK AND NON-CODING RNAS IN PTGR
Non-coding RNAs (ncRNAs) are mRNAs that are not translated into proteins, but are actively transcribed from the human genome. Among many types of ncRNAs, microRNA (miRNA, about 22 nucleotides in length) and long non-coding RNA (lncRNAs, over 200 nucleotides in length) are well-characterized [45]. From the perspective of PTGR, gene expression largely depends on mRNA stability and translation efficiency. RBPs and ncRNAs are well-known trans-acting factors known to play important roles in PTGR by recognizing and interacting with specific RNA sequences termed as cis-elements [46-48]. Even though the oncogenic function of hnRNPK in various cancers has been reported over the past several decades, it is still poorly understood which are the direct RNA targets of hn-RNPK and the mechanism hnRNPK uses to alter the expression of its target genes. Here, we review hnRNPK as an RBP that regulates its direct target RNAs through ncRNAs via PTGR.
HnRNPK and miRNAs
The 3’UTR of mRNAs is a crucial region that determines its stability or translation through trans-acting factors such as RBPs and miRNAs to share consensus binding sequences [49]. Indeed, previous studies showed that miRNA-loaded RNA-induced silencing complex (RISC) complexes and RBPs crosstalk between each other to regulate specific mRNAs cooperatively or competitively [50,51]. Shanmugam et al. [52] reported that cyclooxygenase-2 (COX-2) mRNA, which is induced by the receptor for advanced glycation endproducts (RAGE) ligand S100b, increases its stability through cytoplasmic translocated hnRNPK upon S100b treatment. This group also demonstrated that hnRNPK silencing increases binding affinity for miR-16 in the 3’UTR of COX-2 mRNA. Thus, both hnRNPK and miR-16 competitively regulate COX-2 mRNA stability [52].
We recently demonstrated that hnRNPK silencing inhibits polo like kinase 1 (PLK1) expression in diverse types of cancer cells. More specifically, hnRNPK binds the 3’UTR of PLK1 mRNA in poly r (C) sequences, and interestingly, we identified that the seed region of two miRNAs, miR-149-3p and miR-193b-5p, matches the hnRNPK binding sequence. Mechanistically, hnRNPK overexpression stabilizes PLK1 expression by decreasing the binding affinity of miRNA-loaded RISC complexes in the PLK1 3’UTR C-rich region. Thus, both hnRNPK- and C-rich region-binding miRNAs regulate PLK1 expression competitively [43]. In addition, we reported that prostate tumor overexpressed 1 antisense transcript 1 (PTOV1-AS1) which is directly regulated by hnRNPK, regulates the proto-oncogene heme oxygenase 1 (HMOX1). PTOV1-AS1 has five binding sites for miR-1207-5p and both PTOV1-AS1 and HMOX1 mRNA included the same microRNA response element (MRE) of miR-1270-5p. Therefore, downregulation of PTOV1-AS1 by hnRNPK knockdown inhibits HMOX1 expression through interaction with miR-1207-5p [44].
HnRNPK and lncRNAs
As mentioned above, lncRNAs are RNA transcripts that are longer than 200 nucleotides in length that are not translated into proteins [53,54]. There are several types of lncRNAs, including antisense transcripts, pseudogenes, and long intronic or intergenic RNAs which have recently been reported as functional ncRNAs [55,56]. Some lncRNAs play crucial roles in changing gene expression including chromatin remodeling, translocation, RNA stability, and translation [57]. In addition, several lncRNAs have various emerging roles in cancer progression and development [58]. In this section, we address important roles of lncRNAs with hnRNPK as a regulator for their expression in diverse cancers.
Li et al. [59] very recently identified the cytoplasmic localization of long intergenic non-coding RNA 460 (linc00460), which is up-regulated in non-small cell lung cancer patients compared with normal patients and regulates epithelial-mesenchymal transition (EMT). Through RNA pull-downs and liquid chromatography–mass spectrometry (LC-MS), hnRNPK was shown to interact with linc00460 physically and promote cell migration and invasion [59]. Gong et al. [60] previously reported survival-predictive lincRNA in kidney cancer (SLINKY) as a prognostic marker of clear cell renal cell carcinoma (ccRCC). They identified SLINKY using variable next-generation sequencing data and validated its expression through independent cohorts. The authors also demonstrated that hnRNPK directly binds with SLINKY and those two factors affect cell proliferation and survival in ccRCC [60]. In neuroblastoma, researchers identified the lncRNA Ets-1 promoter-associated non-coding RNA (pancEts-1) as a potential therapeutic target. PancEts-1 is connected with hnRNPK mechanistically and this interaction is stabilized by B-catenin. PancEts-1 is up-regulated in neuroblastoma tissues and promotes cell growth and invasion [61]. Marques Howarth et al. [62] revealed that Ewing sarcoma transcription factor (EWS-FL1) is expressed in primary pediatric human mesenchymal progenitor cells (pMPCs). The authors found that lncRNA-277 (also known as Ewing sarcoma associated transcript 1 [EWSAT1]), interacts with hnRNPK and is increased in EWS-FL1 in pMPCs and repressed target genes. Both EWS-FL1 and EWSAT1 have oncogenic functions; downregulating EWSAT1 diminishes proliferation and colony-forming ability [62]. Translational regulatory lncRNA (treRNA) interacts with hnRNPK and four other RNA binding proteins. treRNA is increased in metastatic breast cancer and controls metastatic potential by regulating a subset of EMT markers [63].
Aberrant expression of Wnt/β-catenin signaling enhances c-Myc and leads to tumorigenesis, especially in colon cancer. Kawasaki et al. [64] examined that the lncRNA termed c-Myc-upregulated lncRNA (MYU), which is a direct target of c-Myc, is associated with hnRNPK and promotes the G1/S transition during the cell cycle to stabilize cyclin dependent kinase 6 (CDK6) in colon cancer cells. Other groups also found that lncRNA cancer susceptibility candidate 11 (CASC11), which is located upstream of c-Myc, is up-regulated in colorectal cancer (CRC) tissues. Furthermore, CASC11 interacts with hnRNPK and positively regulates its expression, thereby affecting Wnt/β-catenin signaling. Therefore, CASC11 is able to regulate metastatic potential in vitro and in vivo via the above pathway [65]. Another example of lncRNA in colon cancer is the Myc-regulated lncRNAs called MYCLos. Kim et al. [66] validated up- or downregulated lncRNAs in CRC tissues through microarray and hnRNPK was shown to be especially involved in the interaction with MYCLo-2. Loss of MYCLo-2 function decreases colon cancer cell proliferation by inducing Myc targets, p21 and p15.
HnRNPK affects not only the regulation of protein-coding mRNAs, but also that of ncRNAs. Through RNA sequencing, we identified and validated that PTOV1-AS1 expression decreased upon hnRNPK silencing and RNP-IP assay showed that PTOV1-AS1 directly interacts with hnRNPK. Similar to the physiological effect of hnRNPK silencing, knockdown of PTOV1-AS1 downregulated cell viability and colony-forming ability. HnRNPK-mediated PTOV1-AS1 regulation modulates HMOX1 expression. In this axis, PTOV1-AS1 has been shown to function as a molecular decoy of miR-1207-5p which is able to bind both PTOV1-AS1 and HMOX1 through the same MRE [44].
CONCLUSIONS
In this review, the roles of hnRNPK in cancer progression through its gene regulatory function were discussed. In particular, we emphasized gene regulation via the interaction of hnRNPK with ncRNAs such as miRNAs and lncRNAs via PTGR. HnRNPK is a highly conserved gene that is abundantly expressed in mammalian cells. Because of the RNA binding motifs and various domains in hnRNPK, it participates in regulating multiple gene processes: transcription, translocation, post-transcription, and translation.
HnRNPK has emerging roles in cancer. Overexpression of hnRNPK induces progression and metastasis in various types of cancer cells. Researchers have demonstrated that high expression of hnRNPK decreases survival rates in cancer patients and silencing or overexpression of hnRNPK controls metastatic potential in vitro and in vivo. Moreover, cytoplasmic accumulation of hnRNPK is related with cancer patient prognosis. Thus, regulation of hnRNPK is strongly suggested to be a promising therapeutic target.
This review focused on hnRNPK-mediated regulation of gene expression at the post-transcriptional level. Both mRNAs and lncRNAs physically interact with hnRNPK and this leads to regulation of RNA stability or translation. In addition, ncRNAs such as miRNAs and lncRNAs are able to cross-talk with hnRNPK to change the expression of their target genes competitively or cooperatively. Furthermore, various cancer-associated lncRNAs and the hnRNPK axis function to regulate the cancer-associated phenotype through modulating target genes. We summarized RNAs that directly bind to hn-RNPK in Table 1.
The target RNAs that are regulated by hnRNPK, either directly or indirectly, are not fully characterized. Although hn-RNPK is a crucial regulator for cancer progression, little is known about the hnRNPK-mediated regulation of targets and its underlying mechanism. To overcome these limitations, high-throughput sequencing data with various approaches will be needed to identify global targets for hnRNPK. Finally, hnRNPK would be a useful bio-marker and may be developed as a future precision medicine target for cancer patients.
Notes
No potential conflict of interest relevant to this article was reported.
Acknowledgements
This work is supported by the National Research Foundation (NRF) of Korea (2017R1A2A2A05069691).